WRFChem¶
WRFChem is numerical weather prediction model coupled with chemistry.
For more information on running WRFChem, see WRFotron.
Simple plot¶
import salem
import matplotlib.pyplot as plt
import matplotlib.gridspec as gridspec
import cartopy.crs as ccrs
ds = salem.open_wrf_dataset(salem.utils.get_demo_file('wrfout_d01.nc'))
ds
<xarray.Dataset>
Dimensions: (bottom_top: 27, south_north: 150, time: 3, west_east: 150)
Coordinates:
lon (south_north, west_east) float32 70.723145 ... 117.81842
lat (south_north, west_east) float32 7.78907 ... 46.457573
* time (time) datetime64[ns] 2008-10-26T12:00:00 ... 2008-10-26T18...
* west_east (west_east) float64 -2.235e+06 -2.205e+06 ... 2.235e+06
* south_north (south_north) float64 -2.235e+06 -2.205e+06 ... 2.235e+06
Dimensions without coordinates: bottom_top
Data variables:
T2 (time, south_north, west_east) float32 301.97656 ... 267.89062
RAINC (time, south_north, west_east) float32 0.0 0.0 0.0 ... 0.0 0.0
RAINNC (time, south_north, west_east) float32 0.0 0.0 0.0 ... 0.0 0.0
U (time, bottom_top, south_north, west_east) float32 ...
V (time, bottom_top, south_north, west_east) float32 ...
PH (time, bottom_top, south_north, west_east) float32 ...
PHB (time, bottom_top, south_north, west_east) float32 ...
T2C (time, south_north, west_east) float32 28.826569 ... -5.259369
PRCP (time, south_north, west_east) float32 nan nan nan ... 0.0 0.0
WS (time, bottom_top, south_north, west_east) float32 ...
GEOPOTENTIAL (time, bottom_top, south_north, west_east) float32 ...
Z (time, bottom_top, south_north, west_east) float32 ...
PRCP_NC (time, south_north, west_east) float32 nan nan nan ... 0.0 0.0
PRCP_C (time, south_north, west_east) float32 nan nan nan ... 0.0 0.0
Attributes:
TITLE: OUTPUT FROM WRF V3.1.1 MODEL
START_DATE: 2008-10-26_12:00:00
SIMULATION_START_DATE: 2008-10-26_12:00:00
WEST-EAST_GRID_DIMENSION: 151
SOUTH-NORTH_GRID_DIMENSION: 151
BOTTOM-TOP_GRID_DIMENSION: 28
DX: 30000.0
DY: 30000.0
GRIDTYPE: C
DIFF_OPT: 1
KM_OPT: 4
DAMP_OPT: 0
DAMPCOEF: 0.01
KHDIF: 0.0
KVDIF: 0.0
MP_PHYSICS: 8
RA_LW_PHYSICS: 1
RA_SW_PHYSICS: 1
SF_SFCLAY_PHYSICS: 2
SF_SURFACE_PHYSICS: 2
BL_PBL_PHYSICS: 2
CU_PHYSICS: 1
SURFACE_INPUT_SOURCE: 1
SST_UPDATE: 0
GRID_FDDA: 0
GFDDA_INTERVAL_M: 0
GFDDA_END_H: 0
GRID_SFDDA: 0
SGFDDA_INTERVAL_M: 0
SGFDDA_END_H: 0
SF_URBAN_PHYSICS: 0
FEEDBACK: 1
SMOOTH_OPTION: 0
SWRAD_SCAT: 1.0
W_DAMPING: 1
MOIST_ADV_OPT: 1
SCALAR_ADV_OPT: 1
TKE_ADV_OPT: 1
DIFF_6TH_OPT: 0
DIFF_6TH_FACTOR: 0.12
OBS_NUDGE_OPT: 0
WEST-EAST_PATCH_START_UNSTAG: 1
WEST-EAST_PATCH_END_UNSTAG: 150
WEST-EAST_PATCH_START_STAG: 1
WEST-EAST_PATCH_END_STAG: 151
SOUTH-NORTH_PATCH_START_UNSTAG: 1
SOUTH-NORTH_PATCH_END_UNSTAG: 150
SOUTH-NORTH_PATCH_START_STAG: 1
SOUTH-NORTH_PATCH_END_STAG: 151
BOTTOM-TOP_PATCH_START_UNSTAG: 1
BOTTOM-TOP_PATCH_END_UNSTAG: 27
BOTTOM-TOP_PATCH_START_STAG: 1
BOTTOM-TOP_PATCH_END_STAG: 28
GRID_ID: 1
PARENT_ID: 1
I_PARENT_START: 1
J_PARENT_START: 1
PARENT_GRID_RATIO: 1
DT: 120.0
CEN_LAT: 29.039997
CEN_LON: 89.79999
TRUELAT1: 29.04
TRUELAT2: 29.04
MOAD_CEN_LAT: 29.039997
STAND_LON: 89.8
POLE_LAT: 90.0
POLE_LON: 0.0
GMT: 12.0
JULYR: 2008
JULDAY: 300
MAP_PROJ: 1
MMINLU: USGS
NUM_LAND_CAT: 24
ISWATER: 16
ISLAKE: -1
ISICE: 24
ISURBAN: 1
ISOILWATER: 14
pyproj_srs: +proj=lcc +lat_0=29.0399971008301 +lon_0...- bottom_top: 27
- south_north: 150
- time: 3
- west_east: 150
- lon(south_north, west_east)float3270.723145 70.97485 ... 117.81842
array([[ 70.723145, 70.97485 , 71.226746, ..., 108.37326 , 108.62515 , 108.87689 ], [ 70.68201 , 70.934265, 71.186676, ..., 108.41333 , 108.66574 , 108.91797 ], [ 70.64075 , 70.89349 , 71.14642 , ..., 108.45355 , 108.70651 , 108.95929 ], ..., [ 61.95575 , 62.31604 , 62.67685 , ..., 116.92316 , 117.283966, 117.64429 ], [ 61.868927, 62.230286, 62.592133, ..., 117.00787 , 117.36975 , 117.73108 ], [ 61.781586, 62.143982, 62.506897, ..., 117.09311 , 117.456024, 117.81842 ]], dtype=float32) - lat(south_north, west_east)float327.78907 7.8294373 ... 46.457573
array([[ 7.78907 , 7.8294373, 7.8693237, ..., 7.8693237, 7.8294373, 7.78907 ], [ 8.038551 , 8.079079 , 8.11908 , ..., 8.11908 , 8.079079 , 8.038551 ], [ 8.288406 , 8.329086 , 8.369247 , ..., 8.369247 , 8.329086 , 8.288406 ], ..., [45.95782 , 46.017597 , 46.076576 , ..., 46.076576 , 46.017597 , 45.95782 ], [46.2079 , 46.267773 , 46.32685 , ..., 46.32685 , 46.267773 , 46.2079 ], [46.457573 , 46.51753 , 46.576714 , ..., 46.576714 , 46.51753 , 46.457573 ]], dtype=float32) - time(time)datetime64[ns]2008-10-26T12:00:00 ... 2008-10-...
array(['2008-10-26T12:00:00.000000000', '2008-10-26T15:00:00.000000000', '2008-10-26T18:00:00.000000000'], dtype='datetime64[ns]') - west_east(west_east)float64-2.235e+06 -2.205e+06 ... 2.235e+06
array([-2235001.48316, -2205001.48316, -2175001.48316, -2145001.48316, -2115001.48316, -2085001.48316, -2055001.48316, -2025001.48316, -1995001.48316, -1965001.48316, -1935001.48316, -1905001.48316, -1875001.48316, -1845001.48316, -1815001.48316, -1785001.48316, -1755001.48316, -1725001.48316, -1695001.48316, -1665001.48316, -1635001.48316, -1605001.48316, -1575001.48316, -1545001.48316, -1515001.48316, -1485001.48316, -1455001.48316, -1425001.48316, -1395001.48316, -1365001.48316, -1335001.48316, -1305001.48316, -1275001.48316, -1245001.48316, -1215001.48316, -1185001.48316, -1155001.48316, -1125001.48316, -1095001.48316, -1065001.48316, -1035001.48316, -1005001.48316, -975001.48316, -945001.48316, -915001.48316, -885001.48316, -855001.48316, -825001.48316, -795001.48316, -765001.48316, -735001.48316, -705001.48316, -675001.48316, -645001.48316, -615001.48316, -585001.48316, -555001.48316, -525001.48316, -495001.48316, -465001.48316, -435001.48316, -405001.48316, -375001.48316, -345001.48316, -315001.48316, -285001.48316, -255001.48316, -225001.48316, -195001.48316, -165001.48316, -135001.48316, -105001.48316, -75001.48316, -45001.48316, -15001.48316, 14998.51684, 44998.51684, 74998.51684, 104998.51684, 134998.51684, 164998.51684, 194998.51684, 224998.51684, 254998.51684, 284998.51684, 314998.51684, 344998.51684, 374998.51684, 404998.51684, 434998.51684, 464998.51684, 494998.51684, 524998.51684, 554998.51684, 584998.51684, 614998.51684, 644998.51684, 674998.51684, 704998.51684, 734998.51684, 764998.51684, 794998.51684, 824998.51684, 854998.51684, 884998.51684, 914998.51684, 944998.51684, 974998.51684, 1004998.51684, 1034998.51684, 1064998.51684, 1094998.51684, 1124998.51684, 1154998.51684, 1184998.51684, 1214998.51684, 1244998.51684, 1274998.51684, 1304998.51684, 1334998.51684, 1364998.51684, 1394998.51684, 1424998.51684, 1454998.51684, 1484998.51684, 1514998.51684, 1544998.51684, 1574998.51684, 1604998.51684, 1634998.51684, 1664998.51684, 1694998.51684, 1724998.51684, 1754998.51684, 1784998.51684, 1814998.51684, 1844998.51684, 1874998.51684, 1904998.51684, 1934998.51684, 1964998.51684, 1994998.51684, 2024998.51684, 2054998.51684, 2084998.51684, 2114998.51684, 2144998.51684, 2174998.51684, 2204998.51684, 2234998.51684]) - south_north(south_north)float64-2.235e+06 -2.205e+06 ... 2.235e+06
array([-2235000., -2205000., -2175000., -2145000., -2115000., -2085000., -2055000., -2025000., -1995000., -1965000., -1935000., -1905000., -1875000., -1845000., -1815000., -1785000., -1755000., -1725000., -1695000., -1665000., -1635000., -1605000., -1575000., -1545000., -1515000., -1485000., -1455000., -1425000., -1395000., -1365000., -1335000., -1305000., -1275000., -1245000., -1215000., -1185000., -1155000., -1125000., -1095000., -1065000., -1035000., -1005000., -975000., -945000., -915000., -885000., -855000., -825000., -795000., -765000., -735000., -705000., -675000., -645000., -615000., -585000., -555000., -525000., -495000., -465000., -435000., -405000., -375000., -345000., -315000., -285000., -255000., -225000., -195000., -165000., -135000., -105000., -75000., -45000., -15000., 15000., 45000., 75000., 105000., 135000., 165000., 195000., 225000., 255000., 285000., 315000., 345000., 375000., 405000., 435000., 465000., 495000., 525000., 555000., 585000., 615000., 645000., 675000., 705000., 735000., 765000., 795000., 825000., 855000., 885000., 915000., 945000., 975000., 1005000., 1035000., 1065000., 1095000., 1125000., 1155000., 1185000., 1215000., 1245000., 1275000., 1305000., 1335000., 1365000., 1395000., 1425000., 1455000., 1485000., 1515000., 1545000., 1575000., 1605000., 1635000., 1665000., 1695000., 1725000., 1755000., 1785000., 1815000., 1845000., 1875000., 1905000., 1935000., 1965000., 1995000., 2025000., 2055000., 2085000., 2115000., 2145000., 2175000., 2205000., 2235000.])
- T2(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- TEMP at 2 M
- units :
- K
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[301.97656, 302.0078 , ..., 301.14062, 301.21875], [301.96094, 301.98438, ..., 301.3203 , 301.39062], ..., [281.4375 , 280.73438, ..., 269.07812, 269.625 ], [281.51562, 280.8672 , ..., 269.3828 , 269.8203 ]], [[301.96875, 301.98438, ..., 301.46875, 301.41406], [301.96875, 302.0625 , ..., 301.5 , 301.4375 ], ..., [279.48438, 279.25 , ..., 268.26562, 268.5547 ], [279.2422 , 278.9453 , ..., 268.65625, 268.71875]], [[301.8672 , 301.875 , ..., 301.40625, 301.28125], [301.875 , 301.95312, ..., 301.375 , 301.2578 ], ..., [278.1875 , 277.625 , ..., 267.6172 , 267.6875 ], [278.14844, 277.60938, ..., 268.07812, 267.89062]]], dtype=float32) - RAINC(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- ACCUMULATED TOTAL CUMULUS PRECIPITATION
- units :
- mm
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.], ..., [0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.]], [[0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.], ..., [0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.]], [[0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.], ..., [0., 0., ..., 0., 0.], [0., 0., ..., 0., 0.]]], dtype=float32) - RAINNC(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- ACCUMULATED TOTAL GRID SCALE PRECIPITATION
- units :
- mm
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.328125, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.695312, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]]], dtype=float32) - U(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- x-wind component
- units :
- m s-1
- stagger :
- X
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- V(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- y-wind component
- units :
- m s-1
- stagger :
- Y
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- PH(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- perturbation geopotential
- units :
- m2 s-2
- stagger :
- Z
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- PHB(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- base-state geopotential
- units :
- m2 s-2
- stagger :
- Z
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- T2C(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- 2m Temperature
- units :
- C
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[28.826569, 28.857819, ..., 27.990631, 28.068756], [28.810944, 28.834381, ..., 28.170319, 28.240631], ..., [ 8.287506, 7.584381, ..., -4.071869, -3.524994], [ 8.365631, 7.717194, ..., -3.767181, -3.329681]], [[28.818756, 28.834381, ..., 28.318756, 28.264069], [28.818756, 28.912506, ..., 28.350006, 28.287506], ..., [ 6.334381, 6.100006, ..., -4.884369, -4.595306], [ 6.092194, 5.795319, ..., -4.493744, -4.431244]], [[28.717194, 28.725006, ..., 28.256256, 28.131256], [28.725006, 28.803131, ..., 28.225006, 28.107819], ..., [ 5.037506, 4.475006, ..., -5.532806, -5.462494], [ 4.998444, 4.459381, ..., -5.071869, -5.259369]]], dtype=float32) - PRCP(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- Total precipitation rate
- units :
- mm h-1
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[ nan, nan, ..., nan, nan], [ nan, nan, ..., nan, nan], ..., [ nan, nan, ..., nan, nan], [ nan, nan, ..., nan, nan]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.109375, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.122396, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]]], dtype=float32) - WS(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- Horizontal wind speed
- units :
- m s-1
- stagger :
- X
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- GEOPOTENTIAL(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- Full model geopotential
- units :
- m2 s-2
- stagger :
- Z
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- Z(time, bottom_top, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XYZ
- description :
- Full model height
- units :
- m
- stagger :
- Z
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
[1822500 values with dtype=float32]
- PRCP_NC(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- Precipitation rate from grid scale physics
- units :
- mm h-1
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[ nan, nan, ..., nan, nan], [ nan, nan, ..., nan, nan], ..., [ nan, nan, ..., nan, nan], [ nan, nan, ..., nan, nan]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.109375, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]], [[0. , 0. , ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ], ..., [0. , 0.122396, ..., 0. , 0. ], [0. , 0. , ..., 0. , 0. ]]], dtype=float32) - PRCP_C(time, south_north, west_east)float32...
- FieldType :
- 104
- MemoryOrder :
- XY
- description :
- Precipitation rate from cumulus physics
- units :
- mm h-1
- stagger :
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
array([[[nan, nan, ..., nan, nan], [nan, nan, ..., nan, nan], ..., [nan, nan, ..., nan, nan], [nan, nan, ..., nan, nan]], [[ 0., 0., ..., 0., 0.], [ 0., 0., ..., 0., 0.], ..., [ 0., 0., ..., 0., 0.], [ 0., 0., ..., 0., 0.]], [[ 0., 0., ..., 0., 0.], [ 0., 0., ..., 0., 0.], ..., [ 0., 0., ..., 0., 0.], [ 0., 0., ..., 0., 0.]]], dtype=float32)
- TITLE :
- OUTPUT FROM WRF V3.1.1 MODEL
- START_DATE :
- 2008-10-26_12:00:00
- SIMULATION_START_DATE :
- 2008-10-26_12:00:00
- WEST-EAST_GRID_DIMENSION :
- 151
- SOUTH-NORTH_GRID_DIMENSION :
- 151
- BOTTOM-TOP_GRID_DIMENSION :
- 28
- DX :
- 30000.0
- DY :
- 30000.0
- GRIDTYPE :
- C
- DIFF_OPT :
- 1
- KM_OPT :
- 4
- DAMP_OPT :
- 0
- DAMPCOEF :
- 0.01
- KHDIF :
- 0.0
- KVDIF :
- 0.0
- MP_PHYSICS :
- 8
- RA_LW_PHYSICS :
- 1
- RA_SW_PHYSICS :
- 1
- SF_SFCLAY_PHYSICS :
- 2
- SF_SURFACE_PHYSICS :
- 2
- BL_PBL_PHYSICS :
- 2
- CU_PHYSICS :
- 1
- SURFACE_INPUT_SOURCE :
- 1
- SST_UPDATE :
- 0
- GRID_FDDA :
- 0
- GFDDA_INTERVAL_M :
- 0
- GFDDA_END_H :
- 0
- GRID_SFDDA :
- 0
- SGFDDA_INTERVAL_M :
- 0
- SGFDDA_END_H :
- 0
- SF_URBAN_PHYSICS :
- 0
- FEEDBACK :
- 1
- SMOOTH_OPTION :
- 0
- SWRAD_SCAT :
- 1.0
- W_DAMPING :
- 1
- MOIST_ADV_OPT :
- 1
- SCALAR_ADV_OPT :
- 1
- TKE_ADV_OPT :
- 1
- DIFF_6TH_OPT :
- 0
- DIFF_6TH_FACTOR :
- 0.12
- OBS_NUDGE_OPT :
- 0
- WEST-EAST_PATCH_START_UNSTAG :
- 1
- WEST-EAST_PATCH_END_UNSTAG :
- 150
- WEST-EAST_PATCH_START_STAG :
- 1
- WEST-EAST_PATCH_END_STAG :
- 151
- SOUTH-NORTH_PATCH_START_UNSTAG :
- 1
- SOUTH-NORTH_PATCH_END_UNSTAG :
- 150
- SOUTH-NORTH_PATCH_START_STAG :
- 1
- SOUTH-NORTH_PATCH_END_STAG :
- 151
- BOTTOM-TOP_PATCH_START_UNSTAG :
- 1
- BOTTOM-TOP_PATCH_END_UNSTAG :
- 27
- BOTTOM-TOP_PATCH_START_STAG :
- 1
- BOTTOM-TOP_PATCH_END_STAG :
- 28
- GRID_ID :
- 1
- PARENT_ID :
- 1
- I_PARENT_START :
- 1
- J_PARENT_START :
- 1
- PARENT_GRID_RATIO :
- 1
- DT :
- 120.0
- CEN_LAT :
- 29.039997
- CEN_LON :
- 89.79999
- TRUELAT1 :
- 29.04
- TRUELAT2 :
- 29.04
- MOAD_CEN_LAT :
- 29.039997
- STAND_LON :
- 89.8
- POLE_LAT :
- 90.0
- POLE_LON :
- 0.0
- GMT :
- 12.0
- JULYR :
- 2008
- JULDAY :
- 300
- MAP_PROJ :
- 1
- MMINLU :
- USGS
- NUM_LAND_CAT :
- 24
- ISWATER :
- 16
- ISLAKE :
- -1
- ISICE :
- 24
- ISURBAN :
- 1
- ISOILWATER :
- 14
- pyproj_srs :
- +proj=lcc +lat_0=29.0399971008301 +lon_0=89.8000030517578 +lat_1=29.0400009155273 +lat_2=29.0400009155273 +x_0=0 +y_0=0 +R=6370000 +units=m +no_defs
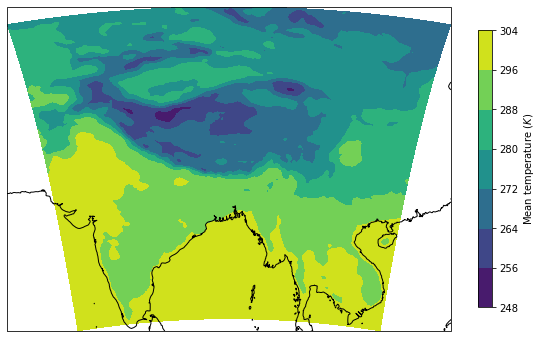
temp_mean = ds['T2'].mean(dim='time')
temp_mean
<xarray.DataArray 'T2' (south_north: 150, west_east: 150)>
array([[301.9375 , 301.95572, 302.00522, ..., 301.41406, 301.33853,
301.3047 ],
[301.9349 , 302. , 302.04688, ..., 301.47134, 301.39844,
301.36197],
[301.9349 , 302.0026 , 302.04166, ..., 301.50522, 301.45572,
301.4297 ],
...,
[279.84634, 279.70053, 279.58594, ..., 267.8047 , 268.3151 ,
268.58072],
[279.70312, 279.20312, 279.33853, ..., 268.08072, 268.3203 ,
268.6224 ],
[279.6354 , 279.14062, 278.6901 , ..., 268.61978, 268.70572,
268.8099 ]], dtype=float32)
Coordinates:
lon (south_north, west_east) float32 70.723145 ... 117.81842
lat (south_north, west_east) float32 7.78907 ... 46.457573
* west_east (west_east) float64 -2.235e+06 -2.205e+06 ... 2.235e+06
* south_north (south_north) float64 -2.235e+06 -2.205e+06 ... 2.235e+06- south_north: 150
- west_east: 150
- 301.9375 301.95572 302.00522 ... 268.61978 268.70572 268.8099
array([[301.9375 , 301.95572, 302.00522, ..., 301.41406, 301.33853, 301.3047 ], [301.9349 , 302. , 302.04688, ..., 301.47134, 301.39844, 301.36197], [301.9349 , 302.0026 , 302.04166, ..., 301.50522, 301.45572, 301.4297 ], ..., [279.84634, 279.70053, 279.58594, ..., 267.8047 , 268.3151 , 268.58072], [279.70312, 279.20312, 279.33853, ..., 268.08072, 268.3203 , 268.6224 ], [279.6354 , 279.14062, 278.6901 , ..., 268.61978, 268.70572, 268.8099 ]], dtype=float32) - lon(south_north, west_east)float3270.723145 70.97485 ... 117.81842
array([[ 70.723145, 70.97485 , 71.226746, ..., 108.37326 , 108.62515 , 108.87689 ], [ 70.68201 , 70.934265, 71.186676, ..., 108.41333 , 108.66574 , 108.91797 ], [ 70.64075 , 70.89349 , 71.14642 , ..., 108.45355 , 108.70651 , 108.95929 ], ..., [ 61.95575 , 62.31604 , 62.67685 , ..., 116.92316 , 117.283966, 117.64429 ], [ 61.868927, 62.230286, 62.592133, ..., 117.00787 , 117.36975 , 117.73108 ], [ 61.781586, 62.143982, 62.506897, ..., 117.09311 , 117.456024, 117.81842 ]], dtype=float32) - lat(south_north, west_east)float327.78907 7.8294373 ... 46.457573
array([[ 7.78907 , 7.8294373, 7.8693237, ..., 7.8693237, 7.8294373, 7.78907 ], [ 8.038551 , 8.079079 , 8.11908 , ..., 8.11908 , 8.079079 , 8.038551 ], [ 8.288406 , 8.329086 , 8.369247 , ..., 8.369247 , 8.329086 , 8.288406 ], ..., [45.95782 , 46.017597 , 46.076576 , ..., 46.076576 , 46.017597 , 45.95782 ], [46.2079 , 46.267773 , 46.32685 , ..., 46.32685 , 46.267773 , 46.2079 ], [46.457573 , 46.51753 , 46.576714 , ..., 46.576714 , 46.51753 , 46.457573 ]], dtype=float32) - west_east(west_east)float64-2.235e+06 -2.205e+06 ... 2.235e+06
array([-2235001.48316, -2205001.48316, -2175001.48316, -2145001.48316, -2115001.48316, -2085001.48316, -2055001.48316, -2025001.48316, -1995001.48316, -1965001.48316, -1935001.48316, -1905001.48316, -1875001.48316, -1845001.48316, -1815001.48316, -1785001.48316, -1755001.48316, -1725001.48316, -1695001.48316, -1665001.48316, -1635001.48316, -1605001.48316, -1575001.48316, -1545001.48316, -1515001.48316, -1485001.48316, -1455001.48316, -1425001.48316, -1395001.48316, -1365001.48316, -1335001.48316, -1305001.48316, -1275001.48316, -1245001.48316, -1215001.48316, -1185001.48316, -1155001.48316, -1125001.48316, -1095001.48316, -1065001.48316, -1035001.48316, -1005001.48316, -975001.48316, -945001.48316, -915001.48316, -885001.48316, -855001.48316, -825001.48316, -795001.48316, -765001.48316, -735001.48316, -705001.48316, -675001.48316, -645001.48316, -615001.48316, -585001.48316, -555001.48316, -525001.48316, -495001.48316, -465001.48316, -435001.48316, -405001.48316, -375001.48316, -345001.48316, -315001.48316, -285001.48316, -255001.48316, -225001.48316, -195001.48316, -165001.48316, -135001.48316, -105001.48316, -75001.48316, -45001.48316, -15001.48316, 14998.51684, 44998.51684, 74998.51684, 104998.51684, 134998.51684, 164998.51684, 194998.51684, 224998.51684, 254998.51684, 284998.51684, 314998.51684, 344998.51684, 374998.51684, 404998.51684, 434998.51684, 464998.51684, 494998.51684, 524998.51684, 554998.51684, 584998.51684, 614998.51684, 644998.51684, 674998.51684, 704998.51684, 734998.51684, 764998.51684, 794998.51684, 824998.51684, 854998.51684, 884998.51684, 914998.51684, 944998.51684, 974998.51684, 1004998.51684, 1034998.51684, 1064998.51684, 1094998.51684, 1124998.51684, 1154998.51684, 1184998.51684, 1214998.51684, 1244998.51684, 1274998.51684, 1304998.51684, 1334998.51684, 1364998.51684, 1394998.51684, 1424998.51684, 1454998.51684, 1484998.51684, 1514998.51684, 1544998.51684, 1574998.51684, 1604998.51684, 1634998.51684, 1664998.51684, 1694998.51684, 1724998.51684, 1754998.51684, 1784998.51684, 1814998.51684, 1844998.51684, 1874998.51684, 1904998.51684, 1934998.51684, 1964998.51684, 1994998.51684, 2024998.51684, 2054998.51684, 2084998.51684, 2114998.51684, 2144998.51684, 2174998.51684, 2204998.51684, 2234998.51684]) - south_north(south_north)float64-2.235e+06 -2.205e+06 ... 2.235e+06
array([-2235000., -2205000., -2175000., -2145000., -2115000., -2085000., -2055000., -2025000., -1995000., -1965000., -1935000., -1905000., -1875000., -1845000., -1815000., -1785000., -1755000., -1725000., -1695000., -1665000., -1635000., -1605000., -1575000., -1545000., -1515000., -1485000., -1455000., -1425000., -1395000., -1365000., -1335000., -1305000., -1275000., -1245000., -1215000., -1185000., -1155000., -1125000., -1095000., -1065000., -1035000., -1005000., -975000., -945000., -915000., -885000., -855000., -825000., -795000., -765000., -735000., -705000., -675000., -645000., -615000., -585000., -555000., -525000., -495000., -465000., -435000., -405000., -375000., -345000., -315000., -285000., -255000., -225000., -195000., -165000., -135000., -105000., -75000., -45000., -15000., 15000., 45000., 75000., 105000., 135000., 165000., 195000., 225000., 255000., 285000., 315000., 345000., 375000., 405000., 435000., 465000., 495000., 525000., 555000., 585000., 615000., 645000., 675000., 705000., 735000., 765000., 795000., 825000., 855000., 885000., 915000., 945000., 975000., 1005000., 1035000., 1065000., 1095000., 1125000., 1155000., 1185000., 1215000., 1245000., 1275000., 1305000., 1335000., 1365000., 1395000., 1425000., 1455000., 1485000., 1515000., 1545000., 1575000., 1605000., 1635000., 1665000., 1695000., 1725000., 1755000., 1785000., 1815000., 1845000., 1875000., 1905000., 1935000., 1965000., 1995000., 2025000., 2055000., 2085000., 2115000., 2145000., 2175000., 2205000., 2235000.])
lon = ds.lon.values
lat = ds.lat.values
fig = plt.figure(1, figsize=(8, 8))
gs = gridspec.GridSpec(1, 1)
ax = fig.add_subplot(gs[0], projection=ccrs.PlateCarree())
ax.coastlines()
im = ax.contourf(lon, lat, temp_mean, transform=ccrs.PlateCarree())
fig.colorbar(im, label='Mean temperature ($K$)', shrink=0.5)
plt.tight_layout()
plt.show()
Problem: Crop arrays to shapefiles¶
Solution 1¶
Customisable cropping
Destaggered/rectilinear grid e.g. after using
pp_concat_regrid.pyon WRFChem output with Salem to destagger and XEMSF to regrid to rectilinear gridFor conservative regridding, consider xgcm.
import numpy as np
import xarray as xr
import geopandas as gpd
from rasterio import features
from affine import Affine
def transform_from_latlon(lat, lon):
""" input 1D array of lat / lon and output an Affine transformation """
lat = np.asarray(lat)
lon = np.asarray(lon)
trans = Affine.translation(lon[0], lat[0])
scale = Affine.scale(lon[1] - lon[0], lat[1] - lat[0])
return trans * scale
def rasterize(shapes, coords, latitude='latitude', longitude='longitude', fill=np.nan, **kwargs):
"""
Description:
Rasterize a list of (geometry, fill_value) tuples onto the given
xarray coordinates. This only works for 1D latitude and longitude
arrays.
Usage:
1. read shapefile to geopandas.GeoDataFrame
`states = gpd.read_file(shp_dir+shp_file)`
2. encode the different shapefiles that capture those lat-lons as different
numbers i.e. 0.0, 1.0 ... and otherwise np.nan
`shapes = (zip(states.geometry, range(len(states))))`
3. Assign this to a new coord in your original xarray.DataArray
`ds['states'] = rasterize(shapes, ds.coords, longitude='X', latitude='Y')`
Arguments:
**kwargs (dict): passed to `rasterio.rasterize` function.
Attributes:
transform (affine.Affine): how to translate from latlon to ...?
raster (numpy.ndarray): use rasterio.features.rasterize fill the values
outside the .shp file with np.nan
spatial_coords (dict): dictionary of {"X":xr.DataArray, "Y":xr.DataArray()}
with "X", "Y" as keys, and xr.DataArray as values
Returns:
(xr.DataArray): DataArray with `values` of nan for points outside shapefile
and coords `Y` = latitude, 'X' = longitude.
"""
transform = transform_from_latlon(coords[latitude], coords[longitude])
out_shape = (len(coords[latitude]), len(coords[longitude]))
raster = features.rasterize(
shapes,
out_shape=out_shape,
fill=fill,
transform=transform,
dtype=float,
**kwargs
)
spatial_coords = {latitude: coords[latitude], longitude: coords[longitude]}
return xr.DataArray(raster, coords=spatial_coords, dims=(latitude, longitude))
# single multipolygon
shapefile = gpd.read_file('/nfs/a68/earlacoa/shapefiles/china/CHN_adm0.shp')
shapes = [(shape, index) for index, shape in enumerate(shapefile.geometry)]
# using regridded file (xemsf)
ds = xr.open_dataset(
'/nfs/b0122/Users/earlacoa/shared/nadia/wrfout_d01_global_0.25deg_2015-06_PM2_5_DRY_nadia.nc'
)['PM2_5_DRY'].mean(dim='time')
# apply shapefile to geometry, default: inside shapefile == 0, outside shapefile == np.nan
ds['shapefile'] = rasterize(shapes, ds.coords, longitude='lon', latitude='lat')
# change to more intuitive labelling of 1 for inside shapefile and np.nan for outside shapefile
# if condition preserve (outside shapefile, as inside defaults to 0), otherwise (1, to mark in shapefile)
ds['shapefile'] = ds.shapefile.where(cond=ds.shapefile!=0, other=1)
# example: crop to shapefile
# if condition (inside shapefile) preserve, otherwise (outside shapefile) remove
ds = ds.where(cond=ds.shapefile==1, other=np.nan) # could scale instead with other=ds*scale
ds.plot()
<matplotlib.collections.QuadMesh at 0x7f3bb67d42b0>

Solution 2¶
Cropping only
Destaggered/rectilinear grid e.g. after using
pp_concat_regrid.pyon WRFChem output with Salem to destagger and XEMSF to regrid to rectilinear gridFor conservative regridding, consider xgcm.
import xarray as xr
import geopandas as gpd
import rioxarray
from shapely.geometry import mapping
shapefile = gpd.read_file('/nfs/a68/earlacoa/shapefiles/china/CHN_adm0.shp', crs="epsg:4326")
ds = xr.open_dataset(
'/nfs/b0122/Users/earlacoa/shared/nadia/wrfout_d01_global_0.25deg_2015-06_PM2_5_DRY_nadia.nc'
)['PM2_5_DRY'].mean(dim='time')
ds.rio.set_spatial_dims(x_dim="lon", y_dim="lat", inplace=True)
ds.rio.write_crs("epsg:4326", inplace=True)
ds_clipped = ds.rio.clip(shapefile.geometry.apply(mapping), shapefile.crs, drop=False)
ds_clipped.plot()
<matplotlib.collections.QuadMesh at 0x7f3bbc9ee280>

Solution 3¶
Staggered grid
WRFChem projection is normally on a Arakawa-C Grid
e.g. intermediate WRFChem files that need to be reused (wrfiobiochemi)
e.g. raw wrfout file (after postprocessing) still on Arakawa-C Grid (2D lat/lon coordinates)
import numpy as np
import xarray as xr
import pandas as pd
import geopandas as gpd # ensure version > 0.8.0
# WARNING: the double for loop of geometry creation and checking is very slow
# the mask in the cell below has already been calculated
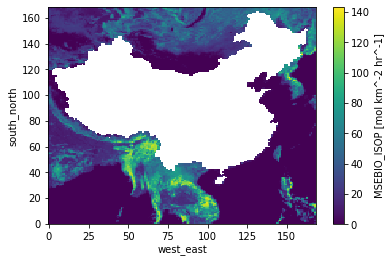
ds = xr.open_dataset('/nfs/b0122/Users/earlacoa/shared/nadia/wrfbiochemi')['MSEBIO_ISOP']
shapefile = gpd.read_file('/nfs/a68/earlacoa/shapefiles/china/CHN_adm0.shp')
mask = np.empty([ds.south_north.shape[0], ds.west_east.shape[0]])
mask[:] = np.nan
for index_lat in range(ds.south_north.shape[0]):
for index_lon in range(ds.west_east.shape[0]):
lat = ds.isel(south_north=index_lat).isel(west_east=index_lon).XLAT.values[0]
lon = ds.isel(south_north=index_lat).isel(west_east=index_lon).XLONG.values[0]
point_df = pd.DataFrame({'longitude': [lon], 'latitude': [lat]})
point_geometry = gpd.points_from_xy(point_df.longitude, point_df.latitude, crs="EPSG:4326")
point_gdf = gpd.GeoDataFrame(point_df, geometry=point_geometry)
point_within_shapefile = point_gdf.within(shapefile)[0]
if point_within_shapefile:
mask[index_lat][index_lon] = True
# bring in mask which computed earlier
ds = xr.open_dataset('/nfs/b0122/Users/earlacoa/shared/nadia/wrfbiochemi')
mask = np.load('/nfs/b0122/Users/earlacoa/shared/nadia/mask_china.npz')['mask']
# demo - removing values in mask
demo = ds['MSEBIO_ISOP'].where(cond=mask!=True, other=np.nan)
demo.plot()
<matplotlib.collections.QuadMesh at 0x7f3bb4be2790>
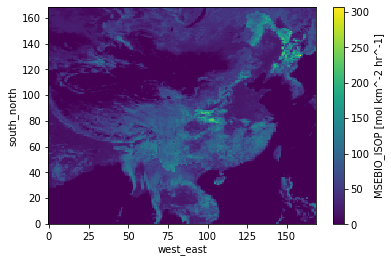
# example - doubling isoprene emissions within mask
ds['MSEBIO_ISOP'] = ds['MSEBIO_ISOP'].where(cond=mask!=True, other=2*ds['MSEBIO_ISOP'])
ds['MSEBIO_ISOP'].plot()
<matplotlib.collections.QuadMesh at 0x7f3bbcd0bbe0>
# saving back into dataset
ds.to_netcdf('/nfs/b0122/Users/earlacoa/shared/nadia/wrfbiochemi_double_isoprene_china')
# check that doubling persisted
ds_original = xr.open_dataset('/nfs/b0122/Users/earlacoa/shared/nadia/wrfbiochemi')['MSEBIO_ISOP']
ds_double = xr.open_dataset('/nfs/b0122/Users/earlacoa/shared/nadia/wrfbiochemi_double_isoprene_china')['MSEBIO_ISOP']
fraction = ds_double / ds_original
print(fraction.max().values)
print(fraction.min().values)
print(fraction.mean().values)
2.0
1.0
1.4608158